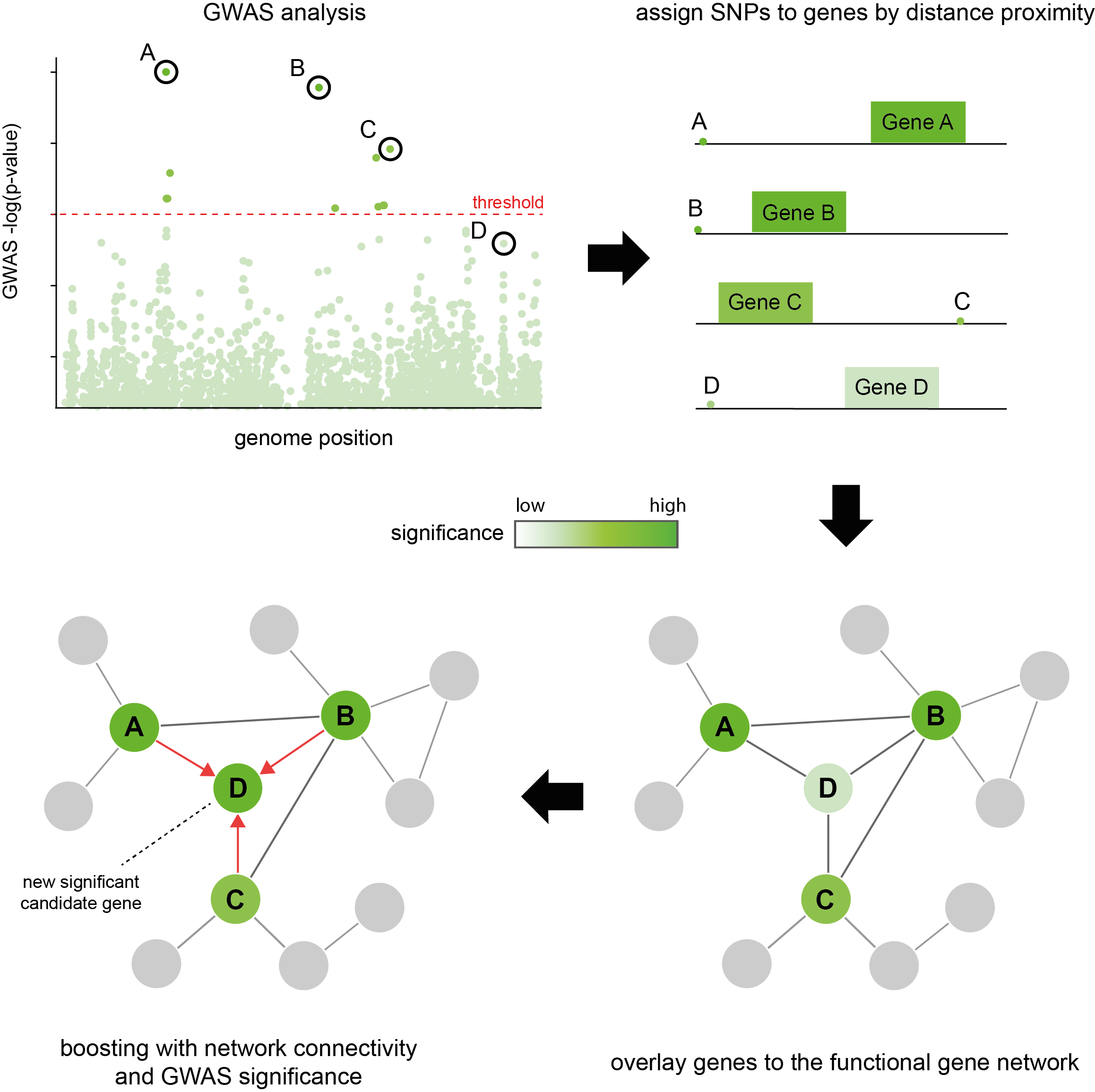
What is araGWAB?
Despite making huge impact on complex trait genetics in humans and plants,
GWAS still suffers from the ‘missing heritability’ in which the identified SNPs cannot explain all the phenotypic variations.
One major reason for missing true phenotype-associated genes is highly strict significance thresholds in GWAS to reduce
false-positives by testing associations for numerous SNPs simultaneously.
The strict P-value threshold after adjustment for multiple hypothesis test such as Bonferroni correction generally
allows only a handful of SNPs to be significant. Presumably,
this statistical limitation may be overcome to some extent by increasing population size, but requiring much higher cost.
araGWAB augments likelihood of association with the given phenotype by integrating GWAS summary statistics (SNP P-values) and co-functional network information.
(1) To use gene-centric significance information, araGWAB first assign SNP P-values to genes based on chromosomal proximity (upper panels)
The genes are assigned to the best P-value of the SNP within user-defined distance (10 kb by default).
Users can also choose the genome build of use, allowing GWAS data based on Affymetrix 250k SNP chip are analyzed on TAIR7 or TAIR8.
Most recent GWAS can be analyzed by selecting TAIR10.
(2) The araGWAB boosts the original GWAS signals using ‘soft’ guilt-by-association (GBA) (lower panels)
with a co-functional network of Arabidopsis genes, AraNet (version 2) (Lee et al., 2015).
A soft GBA approach renders GBA to give full weights only to the genes with a strong GWAS signals.
Assuming the network and GWAS data are conditionally independent, they can be integrated by a naïve Bayesian framework
Major features of araGWAB
araGWAB takes summary statistics (i.e., p-value for SNPs) and known trait-associated genes (derived from disease annotation databases) as user input data, and then reprioritizes genes from GWAS with the following functional features.
- araGWAB internally uses AraNet v2, which includes 84% of the Arabidopsis coding genome.
- araGWAB assigns p-values of SNPs to genes located within a user-defined genomic distance (within 10kbp by default setting) to conduct gene-centric boosting.
- araGWAB allows to analyze GWAS data based on genome builds, TAIR7, TAIR8, and TAIR10.
- araGWAB automatically searches for optimal p-value threshold for boosting within a user-defined range (10-6 < p-value < 10-2), and outputs a list of candidate genes based on the optimal p-value threshold.
- araGWAB completes analysis in an hour for most GWAS set and returns results.
- araGWAB allows users to monitor job status.
- araGWAB reports the boosting results with a summary plot which shows performances (by areas under the ROC curve) of araGWAB, original GWAS data alone, and randomized networks for a given range of p-value threshold.
- araGWAB serves pre-calculated predictions for 9 previous GWAS results.
How to cite araGWAB
araGWAB: Network-based boosting of genome-wide association studies in Arabidopsis thaliana ,Scientific Reports, 2018
Contact information
araGWAB was developed by Lee lab at Yonsei University, Korea.
If you have any question or comment, please contact insuklee(at)yonsei.ac.kr
Updates
2017.09.08 araGWAB web-server launched.