1. Overview of MORPHIN
2. Steps for new query submission
3. Interpretation of the results
4. How to find gene IDs of each model organisms
If you have a question, please contact Sohyun Hwang. [sohyun79(at)yonsei.ac.kr]
1. Overview of MORPHIN
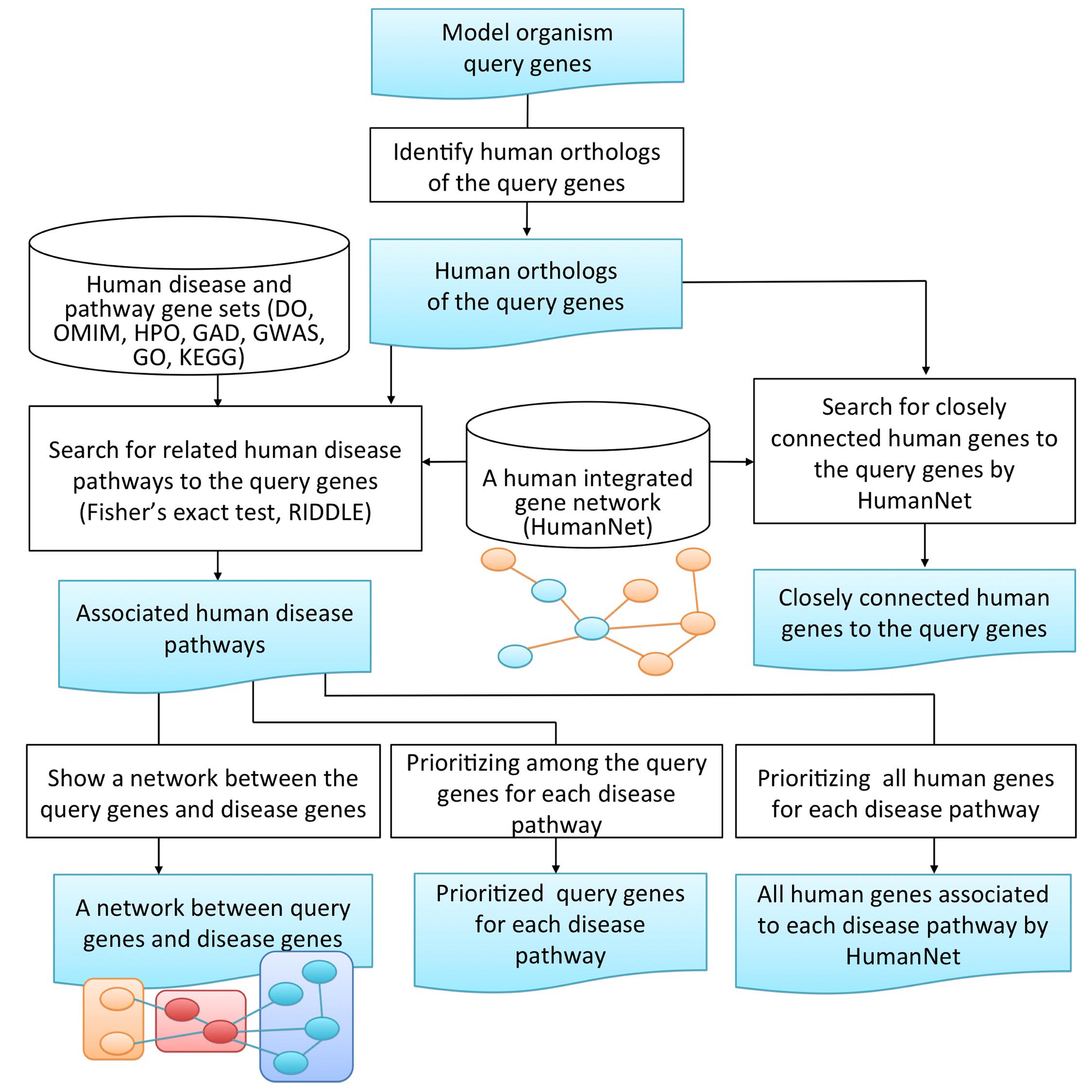
Once a user submits the input query genes of model organism to the MORPHIN query form, the following analytical processes are sequentially performed.
[1] Human ortholog mapping of query genes
MORPHIN entifies human orthologs of the submitted model organism genes using the INPARANOID algorithm.
INPARANOID allows for multiple human orthologs for each model organism gene, where gene expansions have occurred. It uses the pairwise similarity scores calculated by NCBI-Blast between two complete proteomes for constructing orthology groups. An orthology group is initially composed of two-way best hits between two proteomes, and then more sequences are added to the group, if there are sequences in the two proteomes that are closer to the corresponding seed orthologs than to any sequence in the other proteom.
The INPARANOID algorithm achieves a balance between sensitivity and specificity in identifying orthologs across two species by distinguishing in-paralogs duplicated after speciation from out-paralogs duplicated before speciation.
If you need more detailed inforamtion regarding Human ortholog mapping using Inparanoid, Click this.
[2] Search for related human disease pathways to the query genes
MORPHIN identifies significantly related human diseases or pathways (based on associated genes in databases, including OMIM, Disease Ontology, GWAS catalog, Genetic Association Database, GO biological process, Human Phenotype Ontology and KEGG pathway) to the input gene set. Here, MORPHIN uses not only an overlap-based gene set association measure (Fisher exact test) but also a network-based gene set association measure, RIDDLE to enhance the sensitivity of association mapping.
[3] Interpretation of returned search results
MORPHIN returns 6 types of search results.
i) A list of human orthologs for the submitted model organism genes
ii) A list of closely connected genes to human orthologs of query genes in HumanNet
iii) Associated human disease pathways
iv) Networks between human orthologs of query genes and disease genes
v) Prioritized human orthologs of query genes for each disease pathway
vi) Prioritized all human genes for each disease pathway
2. Steps for new query submission
[1] Go to 'Submit New Query' page.
[2] Choose a 'Model species' and an 'Inparalog score threshold'
Currently, MORPHIN takes the following nine model organisms:
| Model organism | Gene names recognized by MORPHIN |
|---|---|
| Saccharomyces cerevisiae (yeast) | ORF ID (e.g., YAL002W) or gene symbol (e.g., VPS8) |
| Caenorhabditis elegans (worm) | Clone ID (e.g., AC3.6) or gene symbol (e.g., col-151) |
| Drosophila melanogaster (fly) | Flybase ID (e.g., FBgn0042137) or gene symbol (e.g., CG18814) |
| Danio rerio (zebrafish) | Entrez ID (e.g., 30590) or gene symbol (e.g., tp53) |
| Mus musculus (mouse) | Entrez ID (e.g., 22059) or gene symbol (e.g., Trp53) |
| Rattus norvegicus (rat) | Entrez ID (e.g., 24842) or gene symbol (e.g., Tp53) |
| Schizosaccharomyces pombe (fission yeast) | Pombase ID (e.g., SPAC22H10.10) or gene symbol (e.g., alp21) |
| Dictyostelium discoideum (soil-living amoeba) | DictyBase ID (e.g., DDB_G0281507) or gene symbol (e.g., colC) |
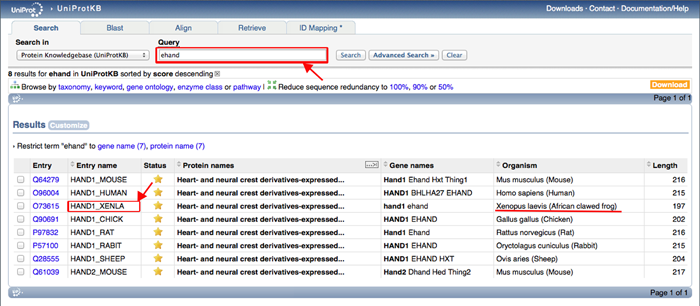
| Xenopus laevis (African clawed frog) | Uniprot Entry name (e.g., HAND1_XENLA) or gene symbol (e.g., ehand) |
Inparalog scores reflects the relative similarity to the two-way best-hit orthologs (from 0 to 1, and 1 indicates maximum likelihood of orthology). We suggest 0 as a default score threshold, which takes all identified orthologs by INPARANOID for MORPHIN analysis. All orthologs identified by INPARANOID are highly reliable, with their statistical significance supported by bootstrapping. However, users can choose scores between 0 (minimal similarity) and 1 (maximal similarity) with higher stringency for orthology.
[3] Choose human disease pathways for MORPHIN analysis
[4] Submit query gene list of the model organism in the text box.
[5] Type an e-mail in text box to receive the search result.
[6] Click 'send' to complete your submission. When your search is done, we will send you the link to see the results by e-mail that you typed.
3. Interpretation of the results
When we search MORPHIN, it shows the following four results. We used, as a toy example, the six worm genes modulating dauer induction: F52D10.3, C54D1.3, Y110A7A.10, F52B5.5, R13H8.1, and F55A3.3
i) A list of human orthologs for the submitted model organism genes

The list of human orthologs for the submitted model organism genes contains information about the model species gene ID and symbol, human ortholog Entrez Gene ID and symbol, Inparanoid score, and Human GO annotations. Inparanoid score is calculated by multiplying two inparalog scores of two genes (model species gene and its human ortholog) provided by Inparanoid algorithm. Inparalog score exists between 1.0 and 0.0. The most confident score is 1.0.
When a model organism gene or symbol has no ortholog or cannot be recognized by MORPHIN ID system, it is printed out in the following separate table. If you want to find a suitable gene ID for unrecognized gene ID or gene symbol, please see 4. How to find gene IDs of each model organisms in Tutorial page.
ii) Closely connected genes to the human orthologs of the query genes in the HumanNet
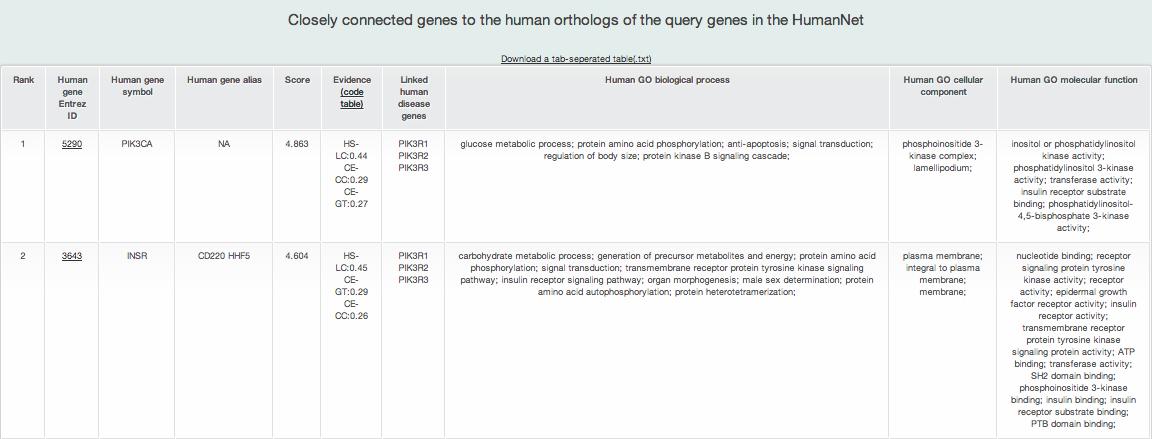
MORPHIN also returns a list of the human genes most strongly linked in HumanNet to the query genes. Such functional associations to the human orthologs of the query genes may provide new biological insights about the function or diseas relationships of the input genes. The result table shows the following seven types of information.
- Rank: the rank on association on the basis of the p-value calculated by Fisher's exact test
- Human Entrez Gene ID
- Human gene symbol
- Human gene alias: gene symbol alias
- Score: prioritization score by a weighted sum of links by HumanNet
- Evidence: supporting the edge score in HumanNet Clicking the code table link shows all HumanNet Evidence codes' description.
- Linked human orthologs of query genes : all human orthologs of query genes connected to the gene
- Three GO annotations
GO biological process: GO biological process annotations for the gene
GO cellular component: GO cellular component annotations for the gene
GO molecular function: GO molecular function annotations for the gene
iii) Associated human disease pathways
MORPHIN uses two algorithms to identify human disease pathways associated to query gene human orthologs. One is Fisher's exact test and the other is RIDDLE. Fisher's exact test is a statistical test measuring significance of the intersection between the query gene human orthologs and the human disease pathway genes. RIDDLE is a network-based method measuring functional closeness between query gene human orthologs and human disease pathway genes. RIDDLE can find relevant disease pathways of query gene human orthologs that are poorly or not at all annotated.
iii-1) Associated human disease pathways by Fisher's exact test
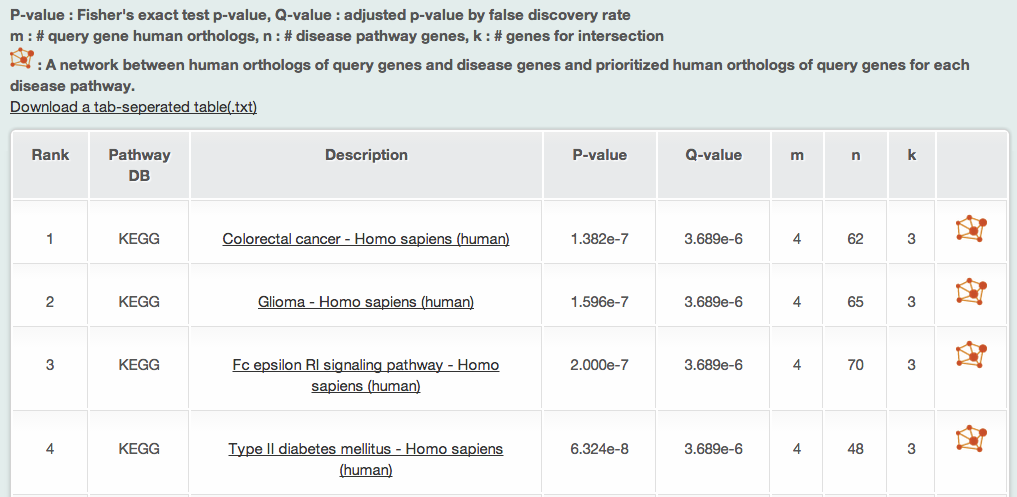
Fisher's exact test analysis returns the eight types of information abut associated human disease pathways to the query gene human orthologs:
- Rank: the rank on association on the basis of the p-value calculated by Fisher's exact test
- Pathway DB
- Pathway description
- p-value: by Fisher's exact test
- q-value: adjusted by false discovery rate
- m: the number of human orthologs of query genes
- n: the number of the associated disease pathway genes
- k: the number of genes common to both the query human orthologs and the disease pathway genes.
Clicking the pathway will show the detailed information of the pathway. However, since the Genetic Association Database and GWAS catalog do not provide the pathway information, MORPHIN does not provide hyperlinks for pathway information.
The above example is the result of query based on the six dauer induction genes. Both Fisher’s exact test and RIDDLE identified diabetes among top ranked human disease pathways. Fisher’s exact test identified 'Type II diabetes mellitus KEGG pathway’ in 4th rank and RIDDLE identified ‘Pregnancy in Diabetes GAD genes' in 3rd rank and 'Type II diabetes mellitus KEGG pathway' in 16th rank. RIDDLE also identified two insulin-related biological processes of 'cellular response to insulin stimulus' and 'insulin-like growth factor receptor signaling pathways' both in 4th ranks.
Clicking the icon of  in the last columns of each disease pathway genes will return a network between human orthologs of query genes and disease genes and prioritized human orthologs of query genes for each disease pathway for each human disease pathways.
in the last columns of each disease pathway genes will return a network between human orthologs of query genes and disease genes and prioritized human orthologs of query genes for each disease pathway for each human disease pathways.
iii-1-a) A network between human orthologs of query genes and Type II diabetes mellitus KEGG pathway identified by Fisher’s exact test
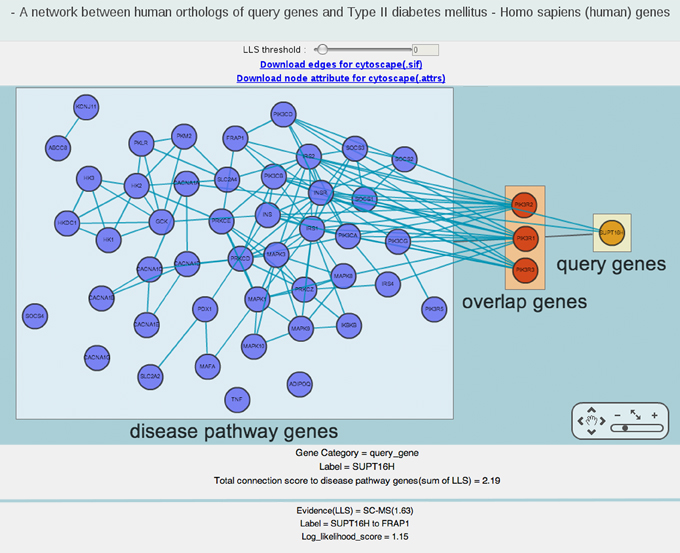
The above network is an example of a network layout view between human orthologs of dauer induction variant genes (i.e., query genes) and genes for Type II diabetes mellitus (i.e., disease pathway genes). Blue, orange, red nodes represent disease pathway genes, human orthologs of query genes and common genes between two gene sets, respectively. If you click a node, the lower panel provides detailed information including the total connection score to disease pathway genes.
Each color-coded nodes are grouped by three boxed areas of query genes, disease pathway genes, and overlap genes. When overlapped common genes exist between query human genes and disease genes, the query gene box is connected to disease gene box through the overlap gene box using black edges.
The green links between two nodes shows the functional relationship based on HumanNet. If you want to see only highly confident links, increase the LLS (log-likelihood scores) threshold in upper panel. The most confident LLS score is 4.26. If you click an edge, the lower panel provides detailed information including log-likelihood score by HumanNet.
You can zoom in and out on the image by clicking the plus and minus buttons, respectively. If you click the hand icon, you can move the view window to other parts of the network.
iii-1-b) Prioritized human orthologs of query genes for Type II diabetes mellitus KEGG pathway identified by Fisher's Exact test
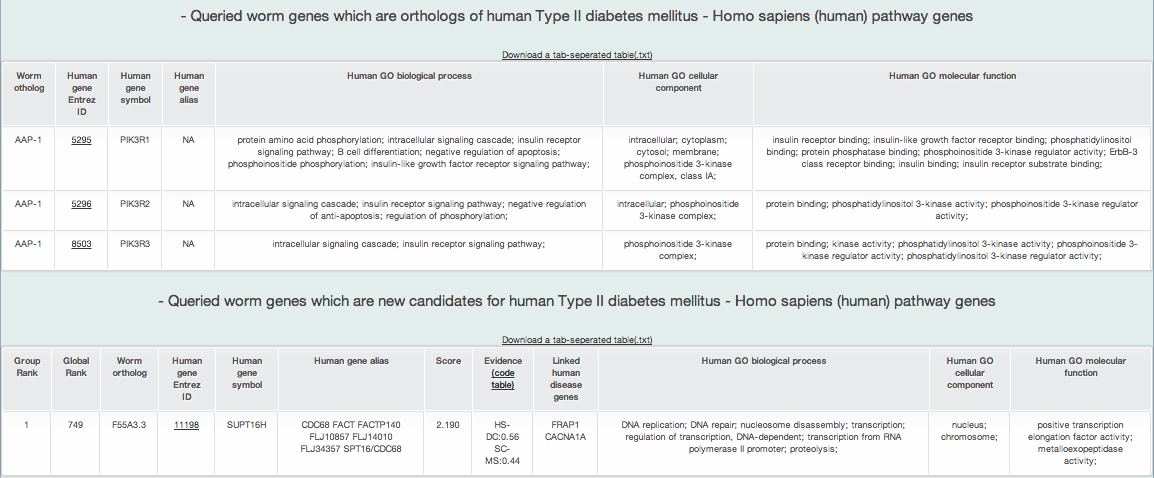
The above two lists are examples of prioritized human orthologs of query genes for KEGG pathway for Type II diabetes mellitus (i.e., the selected disease pathway). MORPHIN first provides a list of human orthologs of query genes overlapping the disease gene set, followed by a list of human orthologs of query genes ranked by their network connectivity scores to the disease gene set.
A new candidate list of human orthologs for the human disease pathway returns the ten types of information :
- Group rank: the rank on the basis of the toal connection score to the disease pathway genes in human orthologs
- Global rank: the rank on the basis of the total connection score to the disease pathway genes in all human genes
- Query gene name: query gene name of model organism
- Human Entrez gene ID: Entrez gene ID of human ortholog of the query gene
- Human gene symbol: gene symbol name of human ortholog
- Human gene alias: gene symbol alias of human ortholog
- Score: prioritization score by the weighted sum of edge weight scores by HumanNet
- Evidence: HumanNet data types (evidence) supporting connections between genes
- Linked_seed: all disease pathway genes connected to the gene
- Three GO annotations
GO biological process: GO biological process annotations for the gene
GO cellular component: GO cellular component annotations for the gene
GO molecular function: GO molecular function annotations for the gene
The evidence of HumanNet data type of each connection between genes are following.
- CE-CC = Co-citation of worm gene
- CE-CX = Co-expression among worm genes
- CE-GT = Worm genetic interactions
- CE-LC = Literature curated worm protein physical interactions
- CE-YH = High-throughput yeast 2-hybrid assays among worm genes
- DM-PI = Fly protein physical interactions
- HS-CC = Co-citation of human genes
- HS-CX = Co-expression among human genes
- HS-DC = Co-occurrence of domains among human proteins
- HS-GN = Gene neighbourhoods of bacterial and archaeal orthologs of human genes
- HS-LC = Literature curated human protein physical interactions
- HS-MS = human protein complexes from affinity purification/mass spectrometry
- HS-PG = Co-inheritance of bacterial and archaeal orthologs of human genes
- HS-YH = High-throughput yeast 2-hybrid assays among human genes
- SC-CC = Co-citation of yeast genes
- SC-CX = Co-expression among yeast genes
- SC-GT = Yeast genetic interactions
- SC-LC = Literature curated yeast protein physical interactions
- SC-MS = Yeast protein complexes from affinity purification/mass spectrometry
- SC-TS = Yeast protein interactions inferred from tertiary structures of complexes
- SC-YH = High-throughput yeast 2-hybrid assays among yeast genes
iii-1-c) Prioritized all human genes for Type II diabetes mellitus KEGG pathway identified by Fisher's Exact test
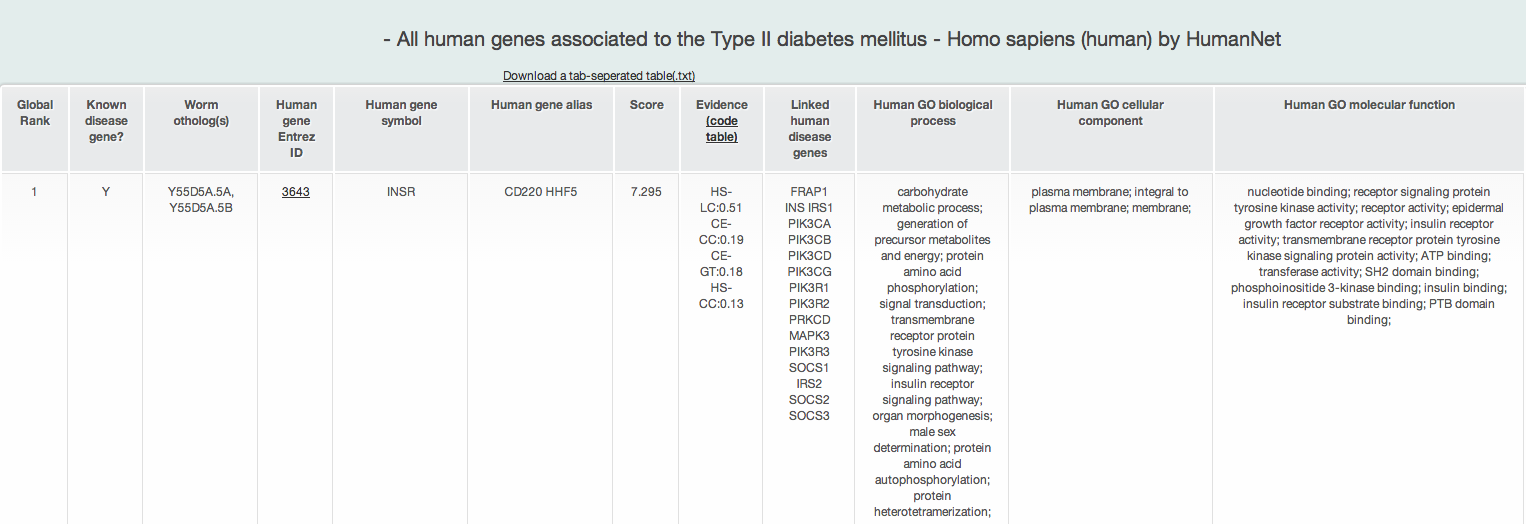
MORPHIN provides a list of prioritized all human genes for each disease pathwyas, as well as a list of prioritized human orthologs of query genes for each disease pathway. However, if you want to see the more information about this prioritization such as its predicitve power, you can use 'Find new members of a pathway' of HumanNet. Search HumanNet with human genes of each disease pathway as a query gene list.
A new candidate list of all human genes for the human disease pathway returns the ten types of information :
- Global rank: the rank on the basis of the total connection score to the disease pathway genes in all human genes
- Known disease gene?: if a gene is known as a disease gene in the disease pathway, it shows Y
- Query gene name: query gene name of model organism
- Human Entrez gene ID: Entrez gene ID of human ortholog of the query gene
- Human gene symbol: gene symbol name of human ortholog
- Human gene alias: gene symbol alias of human ortholog
- Score: prioritization score by the weighted sum of edge weight scores by HumanNet
- Evidence: HumanNet data types (evidence) supporting connections between genes
- Linked_seed: all disease pathway genes connected to the gene
- Three GO annotations
GO biological process: GO biological process annotations for the gene
GO cellular component: GO cellular component annotations for the gene
GO cellular component: GO cellular component annotations for the gene
GO molecular function: GO molecular function annotations for the gene
iii-2) Associated human disease pathways by RIDDLE
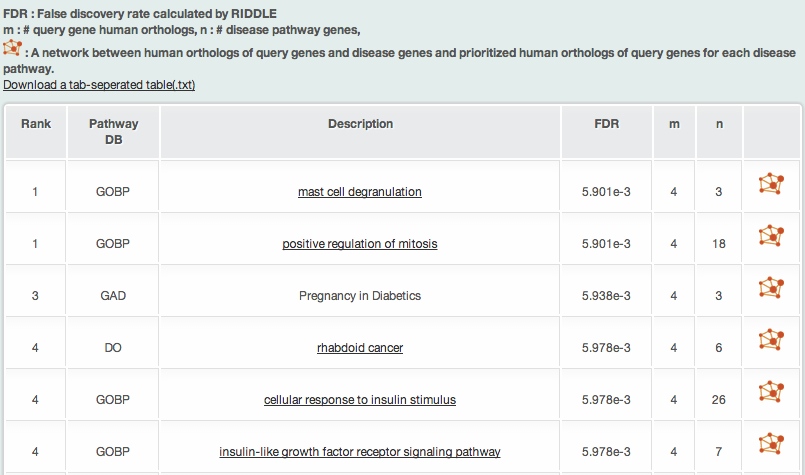
RIDDLE analysis returns the six types of information about associated human disease pathways to the query gene human orthologs:
- Rank: the rank of association on the bases of the FDR (false discovery rate) significance score calculated by RIDDLE
- Pathway DB
- Pathway description
- FDR
- m: the number of query genes
- n: the number of pathway genes.
Clicking the pathway shows the detailed information of the pathway.
The above example is the result of query based on dauer induction genes. Please notice that RIDDLE identified the disease pathway of diabetes that Fisher’s exact test could not identify.
Clicking the icon of  in the last columns of each disease pathway genes, MORPHIN returns a network between human orthologs of query genes and disease genes and prioritized human orthologs of query genes for each disease pathway for each human disease pathways.
in the last columns of each disease pathway genes, MORPHIN returns a network between human orthologs of query genes and disease genes and prioritized human orthologs of query genes for each disease pathway for each human disease pathways.
iii-2-a) A network between human orthologs of query genes and cellular response to insulin stimulus genes identified by RIDDLE
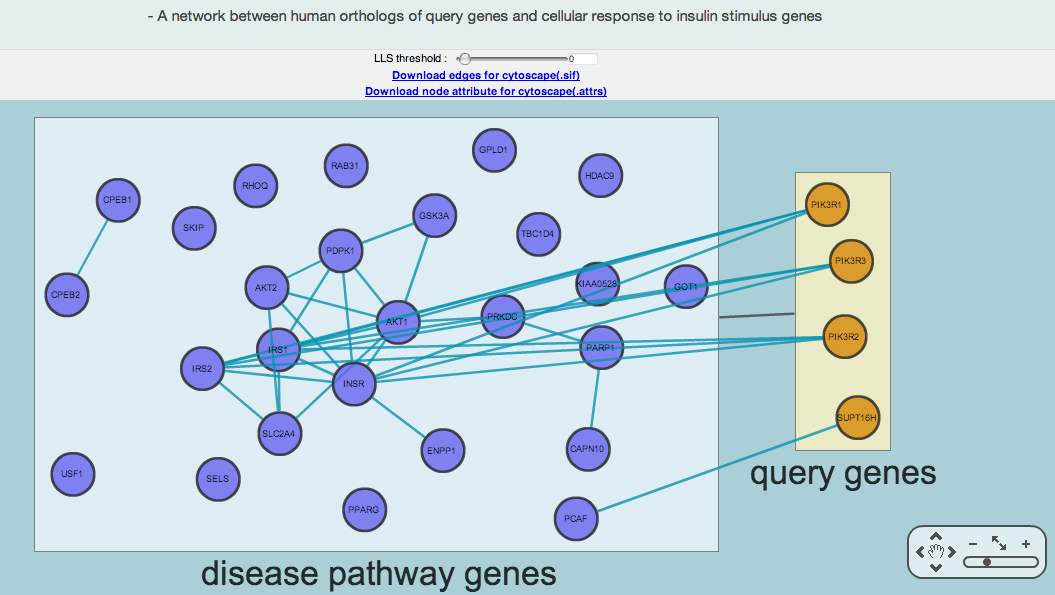
The above network is an example of a network layout view between human orthologs of dauer induction genes (i.e., query genes) and genes for cellular response to insulin stimulus genes (i.e., disease pathway genes). Blue and orange nodes represent disease pathway genes and human orthologs of query genes, respectively.
Since there is no overlapped common gene between query human genes and cellular response to insulin stimulus genes, two boxed areas of query genes and disease pathways genes are connected by a black edge.
The green links between two nodes shows the functional relationship based on HumanNet. If you want to see only highly confident links, increase the LLS (log-likelihood scores) threshold in upper panel. The most confident LLS score is 4.26. If you click an edge, the lower panel provides detailed information including log-likelihood score by HumanNet.
You can zoom in and out on the image by clicking the plus and minus buttons, respectively. If you click the hand icon, you can move the view window to other parts of the network.
MORPHIN shows both group-level connections (black edges) and gene-level connections (blue edges). The RIDDLE algorithm can find connections between a group of query genes and a group of disease genes with no overlapping genes. Sometimes two groups are connected in the absence of gene-level connections between them. This is possible because RIDDLE measures closeness between two groups of genes using not only direct connections but also indirect ones.
iii-2-b) Prioritized human orthologs of query genes for cellular response to insulin stimulus genes identified by RIDDLE
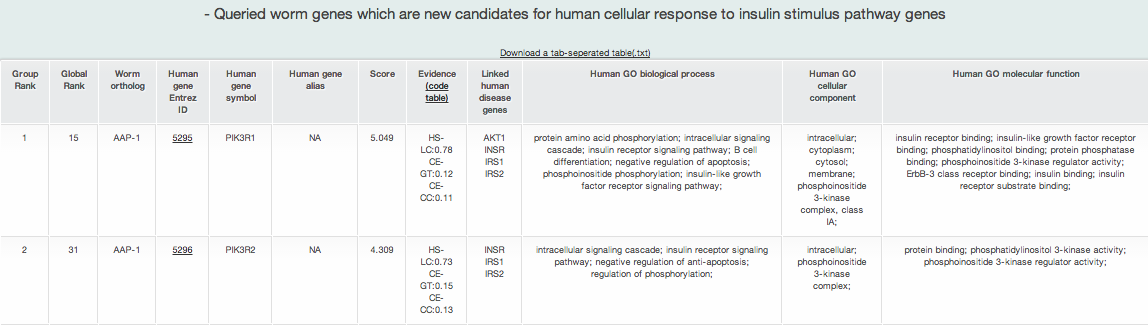
The above list is an example of prioritized human orthologs of query genes for OMIM pathway for cellular response to insulin stimulus (i.e., the selected disease pathway). Among all human orthologs of query genes, it shows only the human orthologs connected to disease pathway genes in HumanNet. It returns the ten types of information about new candidate genes for the disease pathway:
- Group rank: the rank on the basis of the toal connection score to the disease pathway genes in human orthologs
- Global Rank:the rank on the basis of the total connection score to the disease pathway genes in all human genes
- Query gene name: query gene name of model organism Human
- Entrez gene ID: Entrez geneID of human ortholog of a query gene
- Human gene symbol: gene symbol name of human ortholog
- Human gene alias: gene symbol alias of human ortholog
- Score: prioritization score by sum of edge weight scores by HumanNet
- Evidence: HumanNet data types (evidence) supporting connections between genes
- Linked_seed: all disease pathway genes connected to the gene
- Three GO annotations
GO biological process: GO biological process annotations for the gene
GO cellular component: GO cellular component annotations for the gene
GO molecular function: GO molecular function annotations for the gene
The evidence of HumanNet data type of each connection between genes are following.
- CE-CC = Co-citation of worm gene
- CE-CX = Co-expression among worm genes
- CE-GT = Worm genetic interactions
- CE-LC = Literature curated worm protein physical interactions
- CE-YH = High-throughput yeast 2-hybrid assays among worm genes
- DM-PI = Fly protein physical interactions
- HS-CC = Co-citation of human genes
- HS-CX = Co-expression among human genes
- HS-DC = Co-occurrence of domains among human proteins
- HS-GN = Gene neighbourhoods of bacterial and archaeal orthologs of human genes
- HS-LC = Literature curated human protein physical interactions
- HS-MS = human protein complexes from affinity purification/mass spectrometry
- HS-PG = Co-inheritance of bacterial and archaeal orthologs of human genes
- HS-YH = High-throughput yeast 2-hybrid assays among human genes
- SC-CC = Co-citation of yeast genes
- SC-CX = Co-expression among yeast genes
- SC-GT = Yeast genetic interactions
- SC-LC = Literature curated yeast protein physical interactions
- SC-MS = Yeast protein complexes from affinity purification/mass spectrometry
- SC-TS = Yeast protein interactions inferred from tertiary structures of complexes
- SC-YH = High-throughput yeast 2-hybrid assays among yeast genes
iii-2-c) Prioritized all human genes for human cellular response to insulin stimulus pathways genes identified by RIDDLE
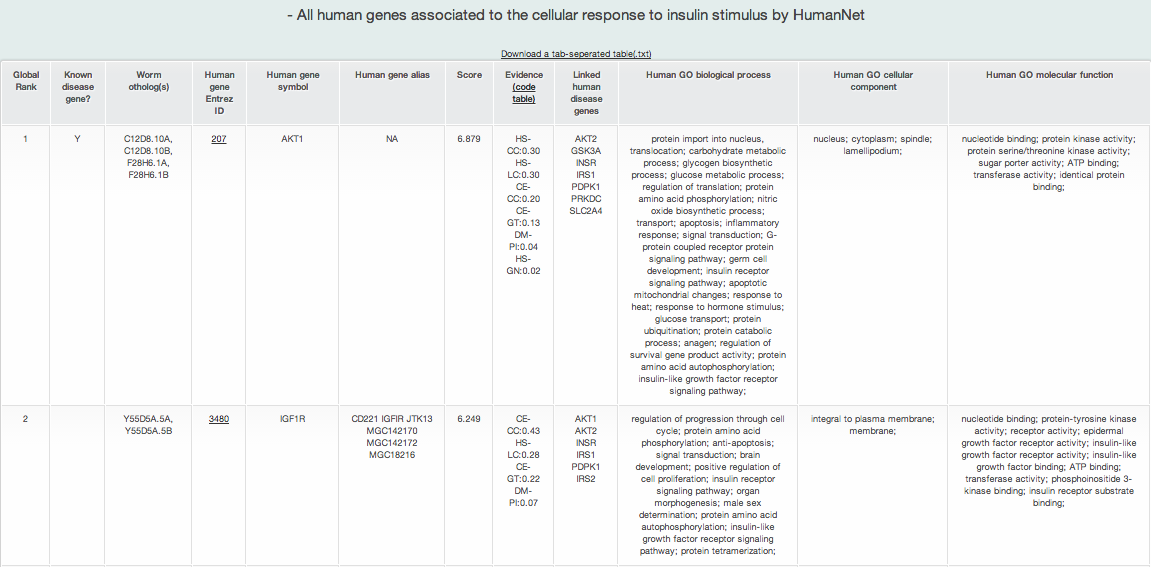
MORPHIN provides a list of prioritized all human genes for each disease pathwyas, as well as a list of prioritized human orthologs of query genes for each disease pathway. However, if you want to see the more information about this prioritization such as its predicitve power, you can use 'Find new members of a pathway' of HumanNet. Search HumanNet with human genes of each disease pathway as a query gene list.
A new candidate list of all human genes for the human disease pathway returns the ten types of information :
- Global rank: the rank on the basis of the total connection score to the disease pathway genes in all human genes
- Known disease gene?: if a gene is known as a disease gene in the disease pathway, it shows Y
- Query gene name: query gene name of model organism
- Human Entrez gene ID: Entrez gene ID of human ortholog of the query gene
- Human gene symbol: gene symbol name of human ortholog
- Human gene alias: gene symbol alias of human ortholog
- Score: prioritization score by the weighted sum of edge weight scores by HumanNet
- Evidence: HumanNet data types (evidence) supporting connections between genes
- Linked_seed: all disease pathway genes connected to the gene
- Three GO annotations
GO biological process: GO biological process annotations for the gene
GO cellular component: GO cellular component annotations for the gene
GO cellular component: GO cellular component annotations for the gene
GO molecular function: GO molecular function annotations for the gene
4. How to find gene IDs of each model organisms
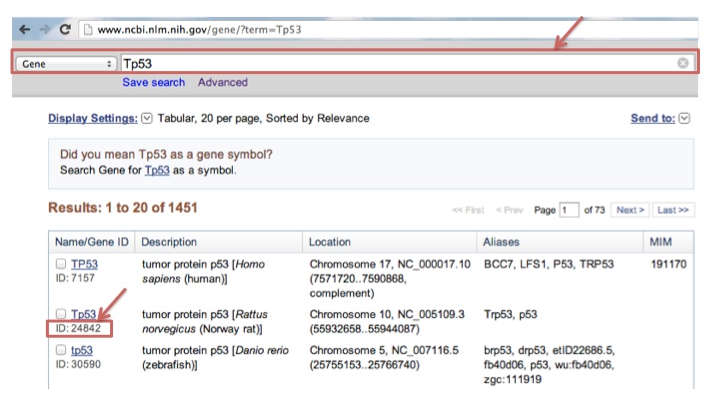
For mouse, rat, zebra fish, MORPHIN uses Entrez Gene ID. To find the Entrez Gene ID of rat Tp53 gene, go to the URL of http://www.ncbi.nlm.nih.gov/gene and submit the gene symbol of Tp53. You can find the Entrez Gene ID 24842 like the following example figure.
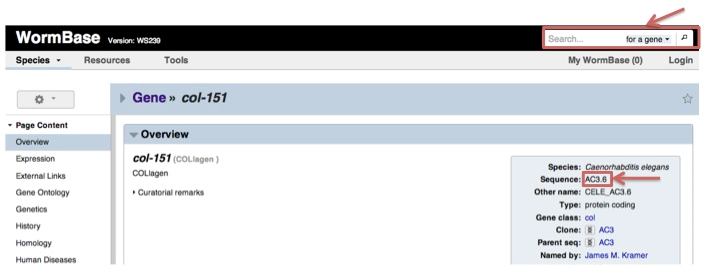
For worm, MORPHIN uses wormbase ID. To find the clone ID of worm col-151 gene, go to the URL of http://www.wormbase.org and submit the gene symbol of col-151. You can find the clone ID (AC3.6) like the following example.
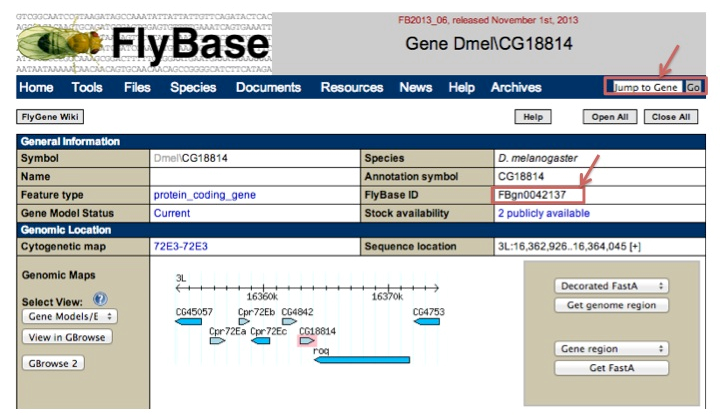
For fly, MORPHIN uses flybase ID. To find the flybase ID of fly CG18814 gene, go to the URL of http://flybase.org and submit the gene symbol of CG18814. You can find the flybase ID (FBgn0042137) like the following example.
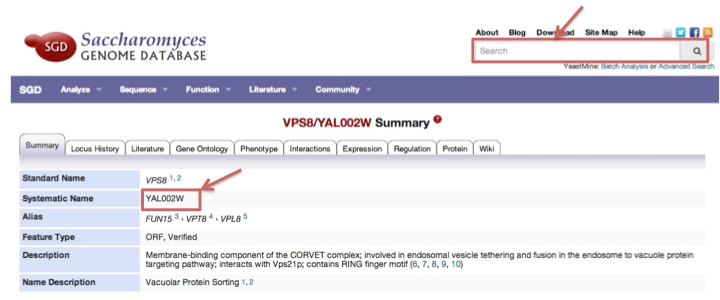
For yeast, MORPHIN uses Saccharomyces genome database ID. To find Saccharomyces ID of yeast VPS8 gene, go to the URL of http://yeastgenome.org and submit the gene symbol of VPS8. You can find the Saccharomyces genome database ID (YAL002W) like the following example.
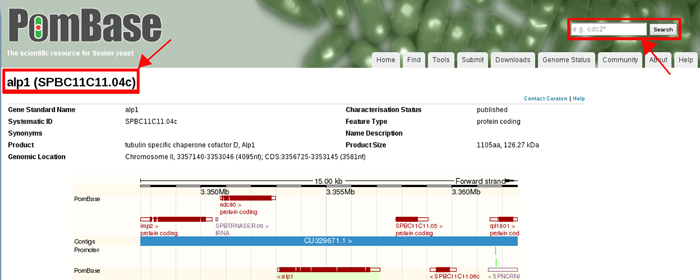
For fission yeast, MORPHIN uses PomBase ID. To find PomBase ID of fission yeast alp21 gene, go to the URL of http://www.pombase.org and submit the gene symbol of alp21. You can find the PomBase ID (SPBC11C11.04c) like the following example.
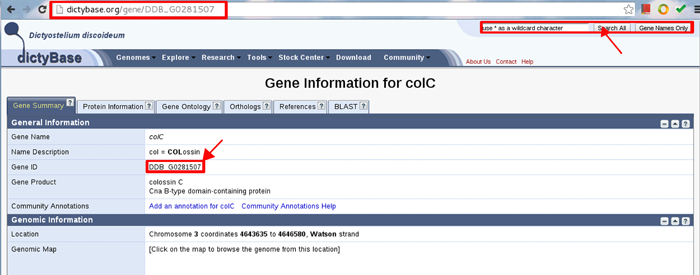
For soil-living amoeba,ORPHIN uses DictyBase ID. To find DictyBase ID of soil-living amoeba colC gene, go to the URL of http://dictybase.org and submit the gene symbol of colC. You can find the DictyBase ID (DDB_GO281507) like the following example.
For African clawed frog, MORPHIN uses UniProt Entry name. To find UniProt Entry name of african clawed frog ehand gene, go to the URL of http://www.uniprot.org/ and submit the gene symbol of ehand. You can find the Uniprot Entry name (HAND1_XENLA) like the following example.